node_classification with GNN
This algorithm is available again, starting with MAGE version 1.22. The algorithm was unavailable in MAGE from version 1.14 to version 1.21.
Node classification is the problem of finding out the right label for a node based on its neighbors’ labels and structure similarities.
The node_classification module supports as follows:
- homogeneous and heterogeneous graphs,
- multiple-label and multi-edge-type graphs,
- any-size datasets,
- the following model architectures:
- Graph Attention with Jumping Knowledge,
- multiple versions of Graph attention networks (GAT),
- GraphSAGE,
- early stopping,
- calculation of various metrics,
- predictions for specified nodes,
- model saving and loading,
- recommendation system use cases.
If you don’t have a vector at the beginning, which will be input for the GNN, then you can call one of the centrality algorithms (PageRank, betwenness centrality, etc.) and save all values into one vector, which will be input for the ML algorithm. That vector usually consists of numbers that describe the dataset in a way that is useful for a particular use case. If that vector is well-defined for the use case, you can expect better results, but if the vector is irrelevant to the use case, you’ll probably get results that are not that useful.
Procedures
This query module contains all necessary functions you need to train GNN model on Memgraph.
The basic node classification workflow is as follows:
- Load data to Memgraph.
- Set parameters by calling the
set_model_parameters()procedure. - Start the training by calling the
train()procedure. - Inspect the training results (optional) by calling
get_training_data()procedures. - Save or load results using the
save_model()andload_model()procedures. - Predict a node class by calling the
predict()procedure.
This MAGE module is still in its early stage. We intend to use it only for exploring or learning about node classification. If you want it to be production-ready, make sure to either open a GitHub issue or drop us a comment on Discord.
You can execute this algorithm on graph projections, subgraphs or portions of the graph.
set_model_parameters()
The set_model_parameters() procedure initializes all global variables. Before
calling the procedure, be sure that node_features property on nodes are in
place.
Input:
-
subgraph: Graph(OPTIONAL) ➡ A specific subgraph, which is an object of type Graph returned by theproject()function, on which the algorithm is run. If subgraph is not specified, the algorithm is computed on the entire graph by default. -
params: (mgp.Map, optional): User defined parameters from query module. Defaults to .
| Name | Type | Default | Description |
|---|---|---|---|
| hidden_features_size | List[Int] | [16, 16] | Embedding dimension for each node in a new layer. |
| layer_type | String | GATJK | Type of layer used, supported types: GATJK, GAT, GRAPHSAGE. |
| aggregator | String | mean | Type of aggregator used, supported type: mean. |
| learning_rate | Float | 0.1 | Optimizer’s learning rate. |
| weight_decay | Float | 5e-4 | Optimizer’s weight decay. |
| split_ratio | Float | 0.8 | Ratio between training and validation data. |
| metrics | List[String] | ["loss","accuracy","f1_score","precision","recall","num_wrong_examples"] | List of metrics to report, supports any combination of “loss”,“accuracy”,“f1_score”,“precision”,“recall”,“num_wrong_examples”. |
| node_id_property | String | id | Property name of node features. |
| num_epochs | Integer | 100 | The number of epochs for model training. |
| console_log_freq | Integer | 5 | Specifies how often results will be printed. |
| checkpoint_freq | Integer | 5 | Specifies how often the model will be saved. The model is persisted on disc. |
| device_type | String | cpu | Defines if the model will be trained using the cpu or cuda. To run on Cuda GPU, check if the system supports it with torch.cuda.is_available(), then set this flag to cuda. |
| path_to_model | String | "/tmp/torch_models" | Path for loading and storing the model. |
Output:
mgp.Record(hidden_features_size=list, layer_type=str, aggregator=str, learning_rate=float, weight_decay=float, split_ratio=float, metrics=mgp.Any, node_id_property=str, num_epochs=int, console_log_freq=int, checkpoint_freq=int, device_type=str, path_to_model=str)➡ Map of parameters set for training
Usage:
When calling the procedure, send only those parameters that need changing from their default values.
CALL node_classification.set_model_parameters(
{layer_type: "GATJK", learning_rate: 0.001, hidden_features_size: [16,16], class_name: "fraud", features_name: "embedding"}
) YIELD aggregator, metrics
RETURN aggregator, metrics;train()
The train() procedure performs model training. Firstly it declares data,
model, optimizer, and criterion. Afterward, it performs training.
Input:
-
subgraph: Graph(OPTIONAL) ➡ A specific subgraph, which is an object of type Graph returned by theproject()function, on which the algorithm is run. If subgraph is not specified, the algorithm is computed on the entire graph by default. -
num_epochs (int, optional)➡ Number of epochs (default:100).
Output:
epoch: int➡ Epoch number.loss: float➡ Loss of model on training data.val_loss: float➡ Loss of model on validation data.train_log: list➡ List of metrics on training data.val_log: list➡ List of metrics on validation data.
Usage:
To train the model, use the following query:
CALL node_classification.train()
YIELD epoch, loss, val_loss
RETURN epoch, loss, val_loss;get_training_data()
Use the get_training_data() procedure to get logged data from training.
Output:
epoch: int➡ Epoch number for current record’s logged data.loss: float➡ Loss in epoch.train_log: mgp.Any➡ Training parameters for epoch.val_log: mgp.Any➡ Validation parameters for epoch.
Usage:
To get logged data, use the following query:
CALL node_classification.get_training_data()
YIELD epoch, loss
RETURN epoch, loss;save_model()
The save_model() procedure saves the model to a specified directory. If there
are already 5 models in the directory, the oldest model is deleted.
Output:
path (str)➡ Path to the stored model.status (str)➡ Status of the stored model.
Usage:
To save the model, use the following query:
CALL node_classification.save_model()
YIELD path, status
RETURN path, status;load_model()
This load_model() procedure loads the model from the specified folder.
Input:
-
subgraph: Graph(OPTIONAL) ➡ A specific subgraph, which is an object of type Graph returned by theproject()function, on which the algorithm is run. If subgraph is not specified, the algorithm is computed on the entire graph by default. -
num (int, optional)➡ Ordinal number of model to load from the default path on the disc (default: 0, i.e., newest model).
Output:
path: str➡ Path of loaded model.
Usage:
To load a model, use the following query:
CALL node_classification.load_model()
YIELD path
RETURN path;predict()
This procedure predicts metrics on one node. It is suggested to load the test data (data without labels) as well. Test data won’t be a part of the training or validation process.
Input:
-
subgraph: Graph(OPTIONAL) ➡ A specific subgraph, which is an object of type Graph returned by theproject()function, on which the algorithm is run. If subgraph is not specified, the algorithm is computed on the entire graph by default. -
vertex: mgp.Vertex➡ Prediction node.
Output:
predicted_class: int➡ Predicted class for specified node.
Usage:
To predict node classification, use the following query:
MATCH (n {id: 1}) CALL node_classification.predict(n)
YIELD predicted_class
RETURN predicted_class;reset()
This function resets all variables to default values.
Output:
status (str)➡ Status of reset function.
Usage:
To reset the values to their default values, use the following query:
CALL node_classification.reset()
YIELD status
RETURN status;Example
Database state
The database contains the following data:
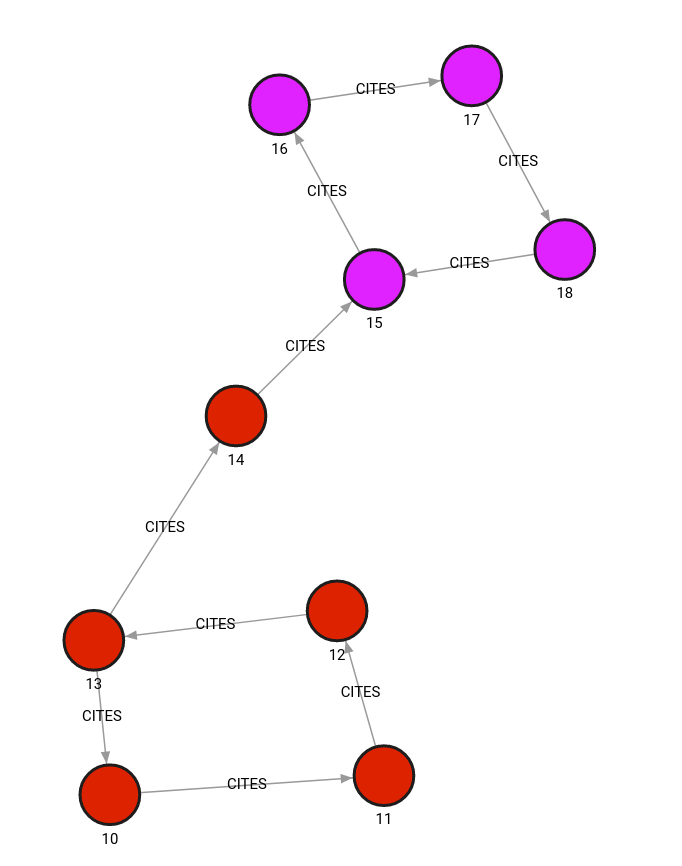
Created with the following Cypher queries:
CREATE (v1:PAPER {id: 10, features: [1, 2, 3], label:0});
CREATE (v2:PAPER {id: 11, features: [1.54, 0.3, 1.78], label:0});
CREATE (v3:PAPER {id: 12, features: [0.5, 1, 4.5], label:0});
CREATE (v4:PAPER {id: 13, features: [0.78, 0.234, 1.2], label:0});
CREATE (v5:PAPER {id: 14, features: [3, 4, 100], label:0});
CREATE (v6:PAPER {id: 15, features: [2.1, 2.2, 2.3], label:1});
CREATE (v7:PAPER {id: 16, features: [2.2, 2.3, 2.4], label:1});
CREATE (v8:PAPER {id: 17, features: [2.3, 2.4, 2.5], label:1});
CREATE (v9:PAPER {id: 18, features: [2.4, 2.5, 2.6], label:1});
MATCH (v1:PAPER {id:10}), (v2:PAPER {id:11}) CREATE (v1)-[e:CITES {}]->(v2);
MATCH (v2:PAPER {id:11}), (v3:PAPER {id:12}) CREATE (v2)-[e:CITES {}]->(v3);
MATCH (v3:PAPER {id:12}), (v4:PAPER {id:13}) CREATE (v3)-[e:CITES {}]->(v4);
MATCH (v4:PAPER {id:13}), (v1:PAPER {id:10}) CREATE (v4)-[e:CITES {}]->(v1);
MATCH (v4:PAPER {id:13}), (v5:PAPER {id:14}) CREATE (v4)-[e:CITES {}]->(v5);
MATCH (v5:PAPER {id:14}), (v6:PAPER {id:15}) CREATE (v5)-[e:CITES {}]->(v6);
MATCH (v6:PAPER {id:15}), (v7:PAPER {id:16}) CREATE (v6)-[e:CITES {}]->(v7);
MATCH (v7:PAPER {id:16}), (v8:PAPER {id:17}) CREATE (v7)-[e:CITES {}]->(v8);
MATCH (v8:PAPER {id:17}), (v9:PAPER {id:18}) CREATE (v8)-[e:CITES {}]->(v9);
MATCH (v9:PAPER {id:18}), (v6:PAPER {id:15}) CREATE (v9)-[e:CITES {}]->(v6);Set model parameters
To set the model parameters, use the following query:
CALL node_classification.set_model_parameters({layer_type: "GAT", learning_rate: 0.001, hidden_features_size: [2,2], class_name: "label", features_name: "features", console_log_freq:1})
YIELD aggregator, metrics
RETURN aggregator, metrics;Train the module
To train the module, use the following query:
CALL node_classification.train(5)
YIELD epoch, loss RETURN *;Results:
+----------+----------+
| epoch | loss |
+----------+----------+
| 1 | 0.788709 |
| 2 | 0.765075 |
| 3 | 0.776351 |
| 4 | 0.727615 |
| 5 | 0.727735 |Predict classification
To predict classification, use the following query:
MATCH (v1:PAPER {id: 10})
CALL node_classification.predict(v1)
YIELD predicted_class
RETURN predicted_class, v1.label as correct_class;Results:
+-----------------+-----------------+
| predicted_class | correct_class |
+-----------------+-----------------+
| 0 | 0 |
+-----------------+-----------------+