
From Documents to Knowledge Graphs: How to Use Unstructured2Graph RAG Tool
Every company sits on a pile of unstructured documents. These can include reports, PDFs, research papers, policies, or meeting notes. They contain valuable knowledge, but little of it is connected or searchable.
With Unstructured2Graph, part of the Memgraph AI Toolkit, you can turn that unstructured text into a connected knowledge graph that LLMs can query and reason over.
Unstructured2Graph combines two powerful components:
- Unstructured IO, which extracts, cleans, and chunks documents of various formats such as PDF, DOCX, or TXT.
- LightRAG, a graph-based reasoning layer that handles prompt engineering and entity extraction automatically, mapping entities and relationships into Memgraph.
Together, they convert raw text into a knowledge graph with nodes, edges, and embeddings ready for retrieval.
In this guide, we’ll walk through how to use the Unstructured2Graph agent step by step using Memgraph Cloud. You’ll quickly go from setting up your project to creating your first entity graph in minutes.
So, let’s get started!
What is Unstructured2Graph RAG Tool
Unstructured2Graph is part of the Memgraph AI Toolkit and serves as an entity extraction and graph creation tool for unstructured data. It automatically converts raw text from documents like PDFs, DOCX, TXT, or HTML, into structured entity graphs inside Memgraph.
It combines the parsing power of Unstructured.io for text extraction and cleaning with the reasoning capabilities of LightRAG for entity recognition and relationship mapping. The result is a queryable entity graph that captures how information connects across your text sources. and is ready in minutes to power your GraphRAG workflows.
Read More: Building Entity Graphs: From Unstructured Text to Graphs in Minutes
Step 1. Sign Up & Create a New Cloud Project
Start by preparing your workspace and running Memgraph:
- Sign up to Memgraph Cloud.
- Create a new project. This will launch a Memgraph instance hosted in the cloud.
- Once it’s ready, copy your connection credentials (host URL, username, and password). You’ll use them to connect from your local environment.
- Open your terminal, VS Code, Cursor, or any other development environment you prefer. This is to run Python scripts connected to your Memgraph Cloud instance.
You are now ready to start building your graph. If you prefer running locally, you can still follow the same steps using Docker, but Memgraph Cloud is the fastest way to begin.
Step 2: Clone the Memgraph AI Toolkit
Next, you need to clone the AI Toolkit repository, which contains the Unstructured2Graph module.
git clone https://github.com/memgraph/ai-toolkit.git
cd ai-toolkit/unstructured2graphStep 3: Install Dependencies
Install uv, the package manager used in the AI Toolkit:
# Install dependencies using uv
uv pip install -e .Details steps are available to follow in its documentation. Once installed, you can use it to run the AI Toolkit packages easily.
Step 4: Connect to Your Cloud Instance
Create a .env file or directly configure your connection to point to the Cloud-hosted Memgraph instance instead of localhost. Example:
MEMGRAPH_URL=bolt+ssc://<your-cloud-instance-ip>:7687
MEMGRAPH_USERNAME=<your_username>
MEMGRAPH_PASSWORD=<your_password>
# Optional for LLM-based entity extraction
OPENAI_API_KEY=your_api_key_hereThis ensures that all extracted entities and relationships will be stored in your Cloud project.
Step 5: Initialize LightRAG & Ingest Documents into Memgraph Cloud
Start by selecting the documents you want to process. Unstructured2Graph supports multiple file types through Unstructured.io, including PDF, DOCX, TXT, and HTML. It extracts readable text, removes unwanted elements such as headers or page numbers, and divides the content into structured chunks based on document layout. Each chunk is then ready for LightRAG to perform entity and relationship extraction.
Next, connect to your Memgraph Cloud instance and initialize the MemgraphLightRAGWrapper. This handles the logic for entity extraction and schema creation.
With this setup, your graph database is ready for ingestion. Keep in mind that LightRAG is stateless. So, if you’ve already run previous imports and want to start clean for a fresh ingestion, you need to clear your LightRAG storage.
Now run the ingestion pipeline to process your unstructured documents directly and load the extracted entities and relationships into your Memgraph Cloud project. You don’t need to define a local DATA_PATH; instead, simply pass your document sources into the pipeline.
import asyncio
from memgraph_toolbox.api.memgraph import Memgraph
from lightrag_memgraph import MemgraphLightRAGWrapper
from unstructured2graph import from_unstructured
async def from_unstructured_with_prep():
memgraph = Memgraph()
memgraph.query("MATCH (n) DETACH DELETE n;")
create_index(memgraph, "Chunk", "hash")
lrag = MemgraphLightRAGWrapper()
await lrag.initialize()
sources = [
"docs/paper.pdf", # local file
"https://example.com/page.html" # remote URL
]
await from_unstructured(
sources=sources,
memgraph=memgraph,
lightrag_wrapper=lrag,
only_chunks=False, # create chunks and extract entities
link_chunks=True # link chunks sequentially with NEXT edges
)
await lrag.afinalize()
if __name__ == "__main__":
logging.basicConfig(level=logging.INFO)
asyncio.run(from_unstructured_with_prep())Here’s what happens step by step:
- Text is extracted, cleaned, and chunked by Unstructured IO.
- Each chunk becomes a
Chunknode in Memgraph with properties likehashandtext. - LightRAG performs entity recognition and relationship extraction, creating
basenodes. - Entities are linked to chunks with
MENTIONED_INedges. - Chunks are connected sequentially with
NEXTedges for traversal.
After processing, your Memgraph instance will hold a complete, queryable knowledge graph.
Step 6: Optimize with indexes and embeddings
To make your knowledge graph ready for semantic retrieval, you first need to compute embeddings and create a vector index. This prepares your data for GraphRAG, where graph-based retrieval happens directly inside Memgraph with a single query.
Here’s how you can do it:
async def full_graphrag(args):
#### INGESTION
memgraph = Memgraph()
if args.ingestion:
await from_unstructured_with_prep(). # This is step 5
compute_embeddings(memgraph, "Chunk")
create_vector_search_index(memgraph, "Chunk", "embedding")
#### RETRIEVAL / GRAPHRAG -> The Native/One-query GraphRAG!
prompt = "What is different under v3.7 compared to v3.6?"
retrieved_chunks = []
for row in memgraph.query(
f"""
CALL embeddings.text(['{prompt}']) YIELD embeddings, success
CALL vector_search.search('vs_name', 5, embeddings[0]) YIELD distance, node, similarity
MATCH (node)-[r*bfs]-(dst:Chunk)
WITH DISTINCT dst, degree(dst) AS degree ORDER BY degree DESC
RETURN dst LIMIT 5;
"""
):
if "description" in row["dst"]:
retrieved_chunks.append(row["dst"]["description"])
if "text" in row["dst"]:
retrieved_chunks.append(row["dst"]["text"])
#### SUMMARIZATION
if not retrieved_chunks:
print("No chunks retrieved. Cannot generate answer.")
else:
context = "\n\n".join(retrieved_chunks)
system_message = prompt_templates.system_message
user_message = prompt_templates.user_message(context, prompt)
if not os.environ.get("OPENAI_API_KEY"):
raise ValueError(
"OPENAI_API_KEY environment variable is not set. Please set your OpenAI API key."
)
client = OpenAI(api_key=os.environ.get("OPENAI_API_KEY"))
completion = client.chat.completions.create(
model="gpt-4o",
messages=[
{"role": "system", "content": system_message},
{"role": "user", "content": user_message},
],
temperature=0.1,
)
answer = completion.choices[0].message.content
print(f"\nQuestion: {prompt}")
print(f"\nAnswer:\n{answer}")from memgraph_toolbox.api.memgraph import Memgraph
from unstructured2graph import compute_embeddings, create_vector_search_index
if __name__ == "__main__":
memgraph = Memgraph()
compute_embeddings(memgraph, "Chunk")
create_vector_search_index(memgraph, "Chunk", "embedding")
#### RETRIEVAL / GRAPHRAG
# The Native/One-query GraphRAG!
for row in memgraph.query(
f"""
CALL embeddings.text(['Hello world prompt']) YIELD embeddings, success
CALL vector_search.search('vs_name', 10, embeddings[0]) YIELD distance, node, similarity
MATCH (node)-[r*bfs]-(dst)
WITH DISTINCT dst, degree(dst) AS degree ORDER BY degree DESC
RETURN dst LIMIT 10;
"""
):
if "description" in row["dst"]:
print(row["dst"]["description"])
if "text" in row["dst"]:
print(row["dst"]["text"])
print("----")
#### SUMMARIZATION
# LLM call with question and retrieved resultsHere’s what happens step by step:
- Embeddings are generated for all
Chunknodes. - A vector index is created to enable fast semantic search.
- A single Cypher query performs the GraphRAG retrieval:
- It converts the input prompt into an embedding.
- It searches for the most semantically relevant chunks.
- It expands context through connected nodes in the graph.
- The retrieved text can then be sent to an LLM for summarization or question answering.
With this, you can perform GraphRAG directly in Memgraph with no separate pipeline required.
Step 7: Visualize the Graph in Memgraph Lab
Open Memgraph Lab and connect to your Cloud instance. Then run:
MATCH (n)-[r]->(m) RETURN n, r, m;You’ll see:
Chunknodes for text sectionsbasenodes for extracted entitiesMENTIONED_INedges linking entities to their source chunksNEXTedges connecting sequential chunks
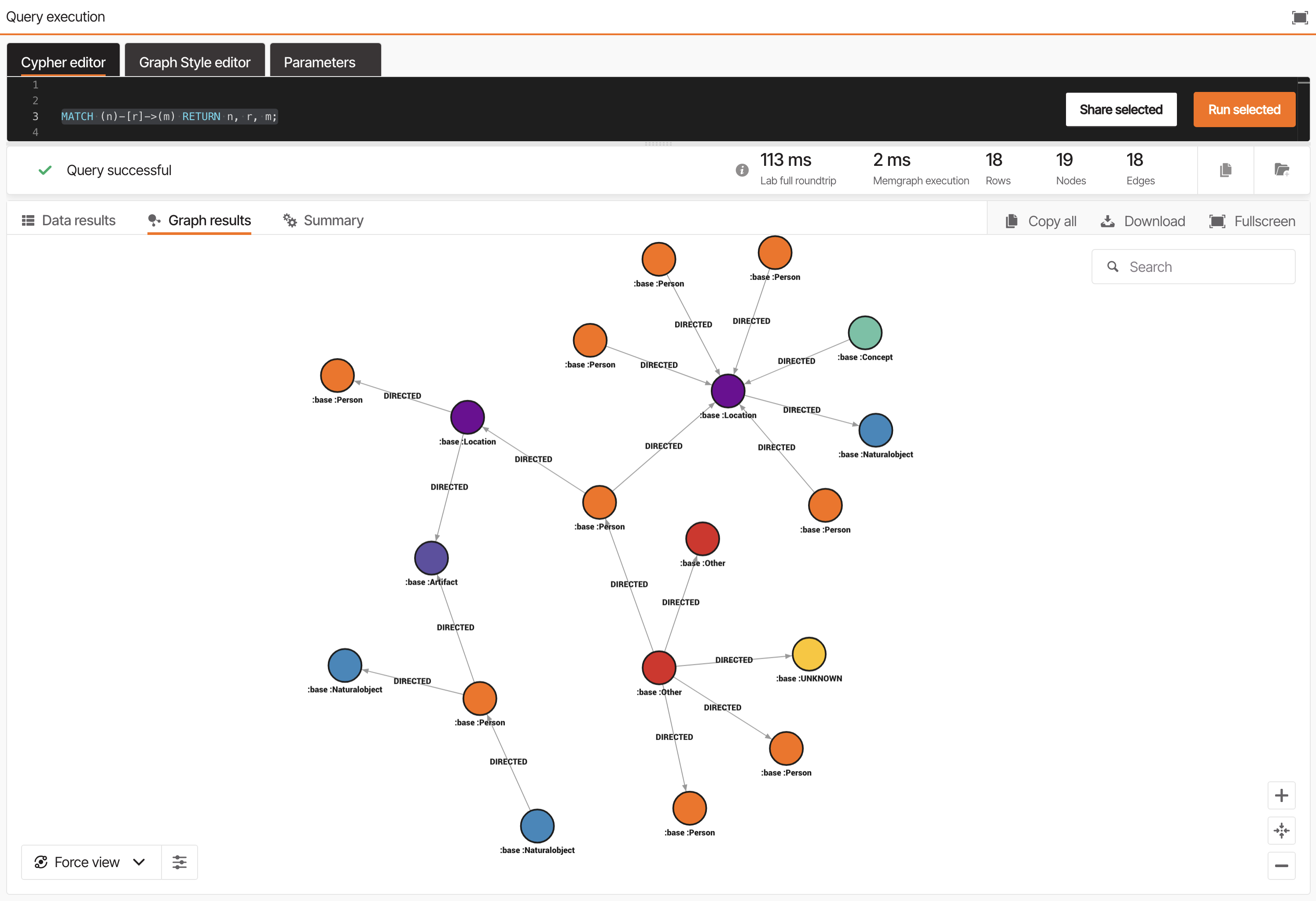
Explore this graph visually to understand how your content has been transformed into a connected network of knowledge.
Memgraph Lab will soon support native multi-agent interaction via MCP Clients, enabling real-time graph reasoning inside the Cloud.
Step 8: Combine With Other Data
Once your document graph is built, you can extend it by linking entities extracted from documents with structured data you’ve already imported through SQL2Graph. This creates a unified knowledge graph that combines structured and unstructured knowledge sources. This is ideal for GraphRAG-powered LLM that requires shared, connected context.
Best Practices for Scaling
Processing unstructured data with LLMs can be resource-intensive, especially across large datasets.
On average, entity extraction may take around 10 seconds per document with ChatGPT 4o Mini.
Plan ingestion in manageable batches and monitor performance using Unstructured2Graph’s built-in time estimates.
Tips for efficiency:
- Start with a small test set to validate quality.
- Adjust chunk size and model choice for better balance between accuracy and speed.
- Clear LightRAG storage between full runs.
- Add vector indexes early for smoother search performance.
Wrapping Up
With just a few lines of Python, you can transform your text documents into a structured graph inside Memgraph. Each paragraph, entity, and relationship becomes part of an interconnected graph that’s ready for querying, visualization, and retrieval-based reasoning.
💡 Try it instantly in Memgraph Cloud
Skip local setup and start using Unstructured2Graph directly in Memgraph Cloud. Sign up, create a new project, and migrate to your instance in just a few minutes.
Alternatively, you can run Memgraph locally via Docker to see how quickly you can go from unstructured text to entity graph.