
Using Memgraph for Knowledge-Driven AutoML in Alzheimer’s Research at Cedars-Sinai
In our recent community call, we had the pleasure of hosting Jason H. Moore, Chair of the Department of Computational Biomedicine at Cedars-Sinai Medical Center, who shared exciting insights into how graph databases are transforming Alzheimer's research.
In the blog post, we’ll unpack how Moore’s team uses Memgraph to build a knowledge-driven AutoML (Automated Machine Learning) pipeline that powers predictive modeling and drug discovery efforts in Alzheimer’s disease.
Watch the full webinar recording – Cedars-Sinai: Using Graph Databases for Knowledge-Aware Automated Machine Learning.
In the meantime, here’s a quick rundown of the key insights from the session.
Talking Point 1: Automated Machine Learning (AutoML) and Alzheimer's Research
Jason H. Moore, a key researcher in Alzheimer's disease, highlighted how AutoML (Automated Machine Learning) is central to his team’s efforts in predicting disease risk and discovering new drug candidates. AutoML automates many steps in the machine learning pipeline, such as model selection and hyperparameter tuning, but it requires high-quality, structured data to function optimally.
Memgraph plays a crucial role here by facilitating the development of a knowledge base that connects multiple, complex biomedical data sources, including patient health records, genetic data, and clinical trial results. These interconnected data points provide the foundation for training AutoML models, resulting in more accurate predictions of disease progression and helping identify potential drug therapies.
Alzheimer’s research is data-heavy and relies on large datasets to extract meaningful insights. Memgraph’s ability to handle this scale and complexity makes it an essential tool in the AutoML pipeline.
Talking Point 2: Memgraph’s Role in Structuring Biomedical Data with KRAGEN and ESCARGOT
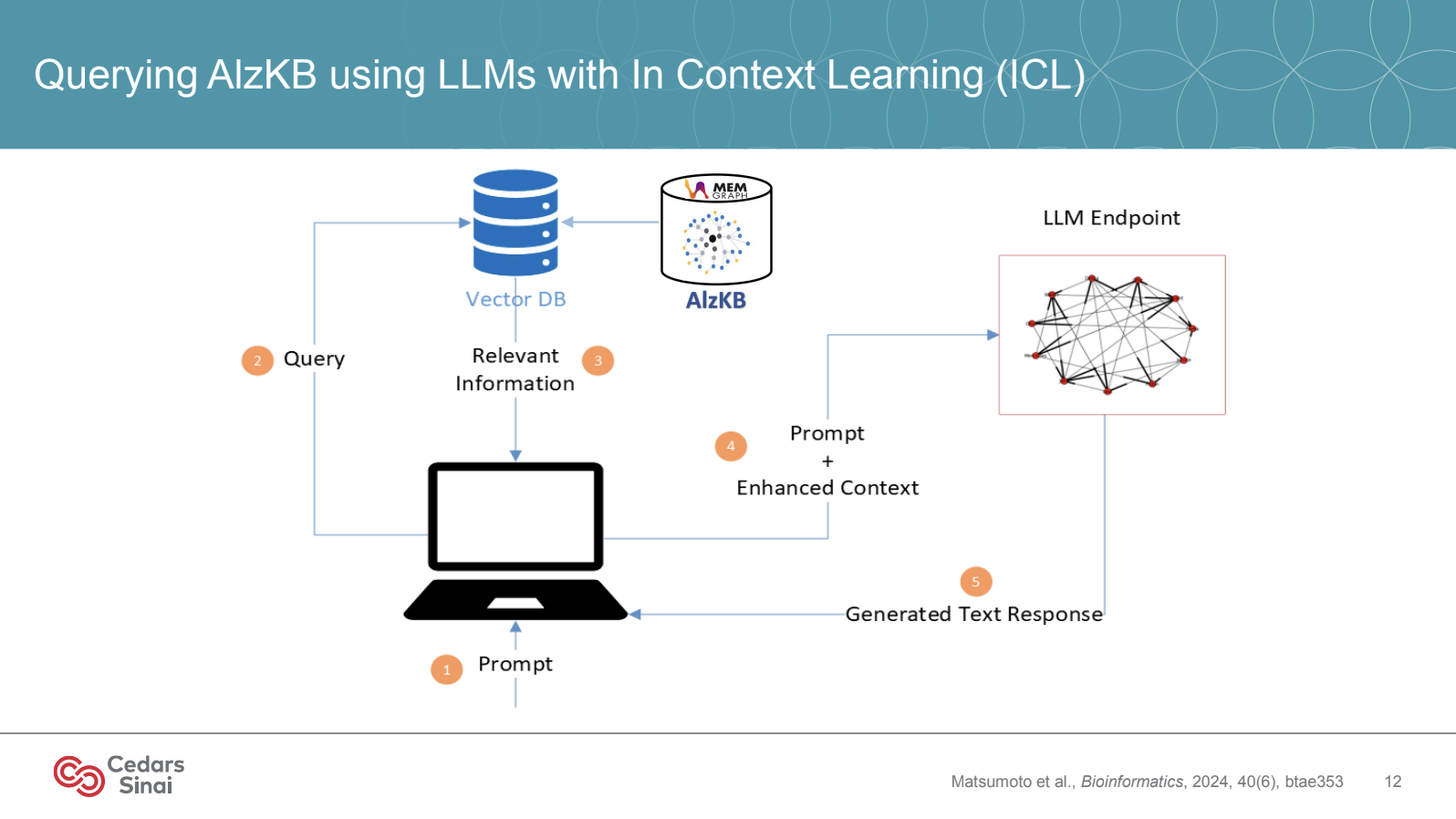
Data is often siloed in biomedical research and challenging to integrate across various sources. Memgraph empowers the team at Cedars-Sinai by supporting a knowledge graph-enhanced RAG (Retrieval-Augmented Generation) framework for biomedical problem-solving with LLMs, using tools like KRAGEN and ESCARGOT.
KRAGEN (Knowledge-Rich AI for Genomics and Environmental Networks) and ESCARGOT are pivotal to how Jason H. Moore’s team structures data within Memgraph. With KRAGEN, the team integrates diverse data sources like genomic data, clinical trial results, and electronic health records (EHRs) into a cohesive knowledge graph, allowing retrieval and exploration of relationships among biological entities—genes, proteins, symptoms, and more. This approach, powered by Memgraph, supports the Graph of Thoughts (GoT), a technique that layers contextual knowledge for LLM-driven biomedical insights, enabling researchers to ask complex queries that would be prohibitively time-consuming in traditional relational databases.
Through Memgraph’s support of KRAGEN and the ESCARGOT system, Moore’s team can instantly visualize and explore relationships within the data. This allows them to uncover and test new hypotheses in Alzheimer’s research, fostering more informed discoveries, particularly around disease progression and intervention strategies.
Talking Point 3: Improving AutoML with Knowledge Graphs
One of the most impactful benefits of using a graph database like Memgraph is how it allows the AutoML pipeline to use contextualized data. The knowledge graph built with Memgraph includes rich, interconnected data that provides context to individual data points, improving the quality and interpretability of machine learning models.
For example, when predicting disease risk, the system can incorporate not just patient demographics and genetic factors but also historical medical conditions, drug interactions, and lifestyle factors—all linked through the graph. This extra layer of contextual information allows the AutoML system to deliver more precise and explainable predictions, which is essential in biomedical research, where understanding why a model makes a specific prediction is critical.
Knowledge graphs also enhance the adaptability of AutoML models, allowing researchers to refine models as new data is continuously integrated.
Talking Point 4: TPOT Integration for AutoML
TPOT, a Python-based tool for AutoML, is used with Memgraph to streamline the machine learning process further. TPOT automates model selection and hyperparameter tuning, which can be time-consuming when done manually.
Memgraph complements TPOT by ensuring the data feeding into the AutoML process is well-structured and connected. This reduces the time spent on data wrangling and preprocessing, enabling researchers to focus on model optimization and evaluation.
This integration of Memgraph with TPOT allows for faster experimentation and quicker insights into drug discovery processes for Alzheimer’s, accelerating the identification of promising therapeutic candidates.
Talking Point 5: Scalability and Ongoing Research
Scalability is a significant requirement in Alzheimer’s research, where datasets grow continuously as new patients are enrolled, new biomarkers are discovered, and new therapies are tested. Memgraph offers the scalability needed to support this ongoing, evolving research.
The knowledge graph is constantly updated with the latest findings from clinical trials, genomic research, and patient outcomes, ensuring that models are always using the most up-to-date data. Memgraph’s ability to handle large, complex datasets makes it the perfect foundation for scaling up both the knowledge base and the machine learning models built on top of it.
As the research expands, Memgraph enables the research team to seamlessly integrate new data sources and continue optimizing their AutoML models without performance degradation, ensuring the continued success of risk prediction and drug discovery efforts.
Q&A
We’ve compiled the questions and answers from the community call Q&A session.
Note that we’ve paraphrased them slightly for brevity. For complete details, watch the entire video.
-
In ESCARGOT, you moved away from using a vector database alone to using both a vector and a graph database. Why did you choose a graph database initially, and what added benefits does it bring when combined with vector embeddings? Specifically, how does the graph structure enhance what you can achieve over a basic RAG setup that relies solely on vector databases?
- Jason: Memgraph's graph database is crucial for this problem because the ontology and graph structure represent the higher-order relationships among all the entities we’re analyzing. This knowledge is critical—and it’s something ChatGPT can’t provide because this specific information isn’t publicly available in a searchable way that a model like ChatGPT would have been trained on. The graph structure allows for a higher-level synthesis of these relationships.
-
Does the ESCARGOT tool support natural language questions that are automatically translated into queries for the knowledge graph?
- Jason: Exactly. The user enters a natural language query, and our AlzKB website will support this soon—within the next few days or early next week. Users with a paid ChatGPT account will be able to log in, enter queries in natural language through the ChatGPT interface, and our KRAGEN and ESCARGOT methods, as described in this session, will process these queries. The system will then retrieve an answer from Memgraph's knowledge graph and deliver it back in natural language, allowing users to interact with the knowledge graph in a straightforward way.
-
How critical is performance for your use case? Do real-time aspects play a significant role for you?
- Jason: Performance has been very good—answers return within seconds. So far, performance hasn’t been an issue. Having Memgraph as an in-memory graph database certainly speeds up the Cypher queries ESCARGOT performs on the fly. Our knowledge graph is relatively small, especially compared to some larger knowledge graphs others may have developed.
-
Does this graph expand over time, or is it mostly static?
- Jason: It will grow over time as we add more entities. While we don’t anticipate adding a large number of new entities, the ones we plan to add will introduce more edges and increase the graph’s size. Even with these additions, the graph will remain well within the memory limits of most computers or laptops used in this application.
-
Can this process be applied to other types of diseases beyond Alzheimer's?
- Jason: Absolutely! We’re about to release an addiction knowledge graph, and we're building one for ALS, another neurodegenerative disease. We have several others in the pipeline as well. The system is open source, so you can easily replace Alzheimer’s data in the knowledge graph with data for any disease you're interested in. It really is almost that straightforward. So, yes, this approach can be specialized for individual diseases or generalized to handle multiple diseases.
-
How critical is having the right ontology for the accuracy of Retrieval-Augmented Generation (RAG)?
-
Jason: That’s a great question. We haven’t specifically tested how critical ontology is to accuracy in answering questions based on the knowledge graph. The questions we see in the table here on the slide are all derived from the graph, so the way the graph is constructed inherently shapes these questions. ESCARGOT or RAG's ability to answer then depends on the methods used.
However, a broader question to ask is: how accurate are the answers we get for Alzheimer’s disease, and how relevant is the knowledge captured by our queries in the context of Alzheimer’s research? For example, does it inform experimental or clinical studies? For this type of question, ontology is indeed very important. It defines the relationships between entities, and the accuracy of these definitions will directly impact the relevance of results to Alzheimer’s disease specifically.
-
-
How rich is the schema of the graph? Are you using multiple properties on both nodes and edges?
- Jason: Right now, our graph is relatively simple. However, we’ve started a project to enhance it by adding more properties to both nodes and edges. This will include features like inferred probabilities for relationships, which we believe will add significant value. We definitely plan to 'color' the graph with additional information to make it richer and more informative. This enhancement is currently in the works, and I think it will contribute a lot to the overall value of the graph.
-
How do you quantify and address uncertainty and inconsistency in knowledge graphs, especially when integrating information from various databases?
- Jason: That’s a great question, and yes, there is inherent uncertainty in each knowledge source we use. The quality of our knowledge graph is only as strong as the sources we’re pulling from and the ontology we’ve constructed, so that uncertainty is always a factor. Personally, I’m not an expert in quantifying that uncertainty, but I agree it’s something we should be addressing. It would be incredibly valuable to communicate to users the degree of uncertainty associated with any relationship in the graph or within the graph as a whole. However, achieving this isn’t straightforward, primarily because we didn’t create the foundational knowledge sources ourselves. We have to rely on their validity, trusting that they’re reliable and well-vetted. Most of our sources have been around for years, are peer-reviewed, published, and widely used in research. While uncertainty exists, we are confident in the quality and reputation of these sources as respected research groups continually update them. So, while it’s not a perfect solution, we can reasonably trust these knowledge sources to a significantly.
-
How do you dynamically update a knowledge graph database and ensure it remains complete?
- Jason: We update our AlzKB knowledge graph at least every few months to align with changes in the underlying knowledge sources. Each time we refresh the graph, we assign a new version number. Our team is dedicated to keeping the graph current, typically with updates on a monthly or bi-monthly basis. Maintaining a continuously updated graph isn’t easy, especially with more frequent updates. However, Memgraph supports dynamic graph updates using triggers and allows data flow in from multiple streams. Memgraph also has online dynamic graph algorithms, which enable the recalculation of values as new data arrives. This setup facilitates a continuously evolving knowledge base, and I believe we’ll see more use cases using dynamic knowledge graphs in the future.
Conclusion
Cedars-Sinai’s Alzheimer’s research demonstrates the power of pairing Memgraph’s graph database with AutoML. By building a scalable, knowledge-rich data structure, Jason H. Moore’s team enhances predictive accuracy and speeds up drug discovery, pushing Alzheimer’s research forward with each new insight.
Watch the full webinar recording – Cedars-Sinai: Using Graph Databases for Knowledge-Aware Automated Machine Learning.
Presentation-Related Links
- https://github.com/epistasislab/TPOT
- https://github.com/epistasislab/KRAGEN
- https://github.com/epistasislab/ESCARGOT
- https://alzkb.ai
Further Reading
- Webinar recording: Cedars-Sinai: Using Graph Databases for Knowledge-Aware Automated Machine Learning
- Blog post: How Precina Health Uses Memgraph and GraphRAG to Revolutionize Type 2 Diabetes Care with Real-Time Insights
- Webinar recording: Optimizing Insulin Management: The Role of GraphRAG in Patient Care
- Blog post: Building GenAI Applications with Memgraph: Easy Integration with GPT and Llama
- Blog post: How Microchip Uses Memgraph’s Knowledge Graphs to Optimize LLM Chatbots
- Webinar recording: Microchip Optimizes LLM Chatbot with RAG and a Knowledge Graph
- User story: Enhancing LLM Chatbot Efficiency with GraphRAG (GenAI/LLMs)
Memgraph Academy
If you are new to the GraphRAG scene, check out a few short and easy-to-follow lessons from our subject matter experts. For free. Start with: